Noncoding RNA, the case of the LC ribozyme
All the functions of the body are registered within the genes of each individual. There are even parts of genes called "introns" which, theoretically, code nothing at all. These portions of genes are normally removed during transcription, but not always. The collaboration of several European teams have made it possible to better understand the mechanisms allowing certain introns to actively react during the stage known as "splicing", and to regulate translation.
Before being translated into proteins by ribosomes, the genes within living organisms must be transcribed into messenger RNA. The coding sequence of the messengers must be continuous to be correctly read by the particles producing proteins, the ribosomes. However, the primary transcripts present an alternation of sequences coding protein fragments (exons) and noncoding sequences (introns). These introns must be eliminated by a splicing process whose various mechanisms rely on catalytic RNA, also termed ribozymes.
Mainly in yeasts, fungi and plants, sequences of catalytic introns are integrated into the primary transcripts. These introns are regarded as parasitic sequences since they can be disseminated in the genomes and transmitted between species based on specific mechanisms. They often come with sequences coding for maturation protein which facilitate their dissemination. For a long time, scientists believed that the presence of such introns in the primary transcripts was neutral for the host body. Many observations, however, now show that it is not the case.
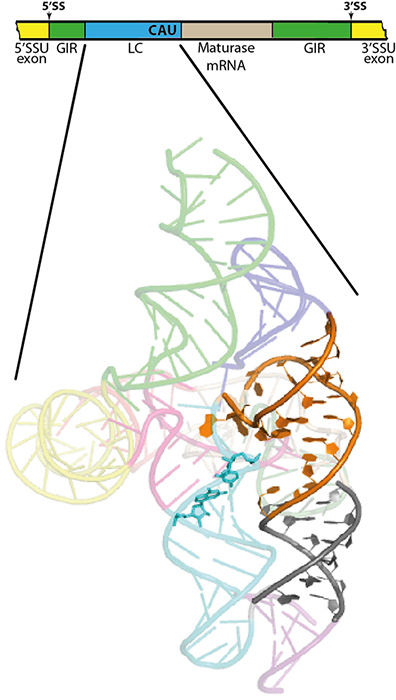
It is in this context that the discoveries of a European team take place. Part of the team started by identifying autocatalytic introns which are accompanied by an additional ribozyme forming a small loop of three nucleotides at the end of the messenger of the intron-coded maturase. This lariat-shaped cap (hence the name of LC ribozyme) protects the maturase messenger from degradation and thus provides the intron with a selective advantage by increasing its chances of dissemination. Until now, LC ribozyme has always been found in a primary transcript of ribosomal RNA. The activities of both ribozymes must thus be coordinated to avoid the inopportune cut of the primary transcript which would prevent the formation of ribosomes and thus the translation.
Benoit Masquida, joint author of the study and researcher at GMGM (Molecular Genetics Genomics and Microbiology, a joint research unit involving Strasbourg University and CNRS) tells us more about the experiments undertaken on two synchrotrons: "Through the crystal structures and small-angle X-ray scattering solved thanks to the data obtained at the Paul Scherrer Institute-Swiss Light Source (x06da) and the SOLEIL synchrotron (PROXIMA 1 and SWING beamlines), we were able to better understand the mechanism allowing the activation of LC ribozyme as well as its catalytic mechanism. Moreover, we were able to establish that the LC ribozyme derives from an autocatalytic intron which would have fit within another intron where it underwent a speciation process generating structures specialized in the control of a new catalytic activity.”