Tracking the accumulation of antibiotic molecules in resistant bacteria
The Joint Research Unit UMR-MD1, Aix-Marseille University, has been working with the DISCO beamline for several years to develop a non-invasive, non-perturbing method of determining the concentration and location of common antibiotics or new antibiotic molecules in bacteria (see also http://www.synchrotron-soleil.fr/Soleil/ToutesActualites/2010/BacteriesResistantes). New results have just been published.
The team led by Jean-Marie Pagès took an ambitious approach, aiming to examine both the bacterial population as a whole and the isolated bacterium within this population.
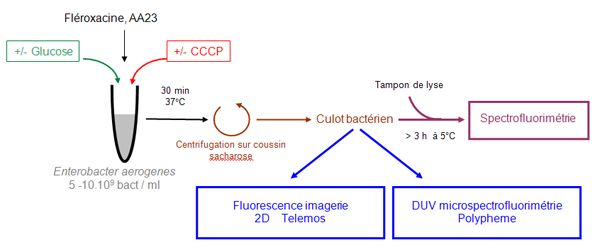
The method developed on the DISCO beamline is based on the simultaneous analysis of a sample via fluorimetry, microfluorimetry, and 2D imaging, as presented in the diagram in Figure 1.
This combined experimental approach obtains the concentration of the molecule being studied on the scale of the population (macro-analysis), on the scale of the isolated bacterium (micro-analysis), and the 2D location on an isolated bacterium (Ref. 1-2).
Along with this approach, the antibacterial activity is determined by measurement on the same strains, to correlate the activity, location, and concentration of the antibiotics being studied. The measurement is taken on strains expressing or not expressing the efflux system responsible for the expulsion of the antibiotics (Ref. 3).
'Old' and new antibiotics
The antibiotics tested are fleroxacin, an antibiotic used for many years, to which bacteria have developed resistance mechanisms, and AA23, an original molecule produced recently by the Isabelle Artaud’s team (Ref. 3).
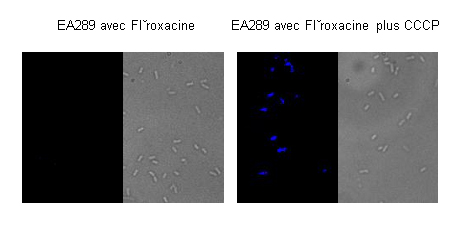
Figure 2A shows that the accumulation of the antibiotic in the bacteria cannot be seen in the presence of fleroxacin alone, because the fleroxacin (fluorescent) does not remain inside the bacterial cell; the bacteria expel it using their efflux system, which is involved in the resistance mechanism that contribute to multidrug resistance phenotype. If an energy decoupler (CCCP) is added, it disturbs the efflux activity, so that the antibiotic remains in the bacteria. We then see bacterial fluorescence, which indicates an accumulation of fleroxacin in those bacteria.
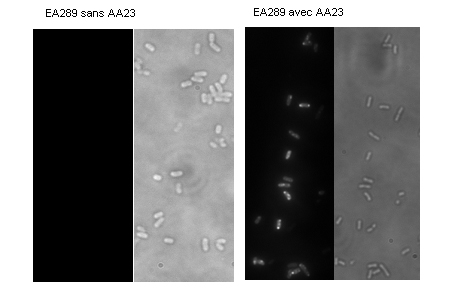
The AA23 molecule was designed to circumvent this efflux system. This is shown in Figure 2B: with the molecule alone, without the addition of CCCP, we already obtain a marking indicating that AA23 is accumulating in the bacteria. We observe that this marking occurs at certain specific locations in the bacteria.
Interestingly, although the addition of the energy decoupler restores the accumulation of fleroxacin in the resistant bacteria, we see a heterogeneity in the fluorescence labelling, which reflects a difference in behaviour on the scale of the isolated bacterium belonging to the population treated in this way. This result illustrates the bases of bacterial individualism and/or the flexibility of adaptation in the presence of environmental signals such as antibacterial agents.
References:
1 - Kaščáková S et al. “Antibiotic Transport in Resistant Bacteria: Synchrotron UV Fluorescence Microscopy to Determine Antibiotic Accumulation with Single Cell Resolution.” PLoS One, 2012, 7(6): art; n° 38624
2 - Pagès J.M. et al. "New peptide-based antimicrobials for tackling drug resistance in bacteria: single cell fluorescence imaging.” Med. Chem. Letters, 2013, 4:556.
3 - Nikaido H. & Pagès J.M. “Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria.” FEMS Microbiol Rev. 2012, 36:340.