A new context for virtual histology: 3D X-ray microimaging of the brain during paraffin embedding
Microscopic imaging of the brain to identify diseases has traditionally relied on histology, where the brain tissue is embedded into paraffin wax and thinly sliced for investigation under a microscope. A disadvantage of this method is that it only provides 2D information and the preparation of the samples causes non-uniform shrinkage of the brain tissue. Using 3D X-ray phase-contrast microtomography, an interdisciplinary team of researchers have now identified these distortions and determined which methods of preparation are best suited to obtaining contrast-rich 3D X-ray images of brain tissue.
Histology and histopathology, i.e., the science of tissue structure and of the microscopic structure of diseased tissue, are the standard tools both in clinical practice – for example to diagnose cancer – and in medical research, including neuroimaging. These methods rely on thin sectioning of tissue prepared specifically for examination under a microscope. The preparation of the samples involves a lengthy procedure of chemical fixation, dehydration, embedding, and usually staining, which alters the chemical and physical properties of the sample tissue including the shape and size of anatomical features. Further, conventional histology only yields two-dimensional images, usually distorted due to strain during slicing. High-resolution 3D X-ray microtomography has earned the name virtual histology, as this sectioning-free imaging technique can overcome certain limitations of traditional histology.
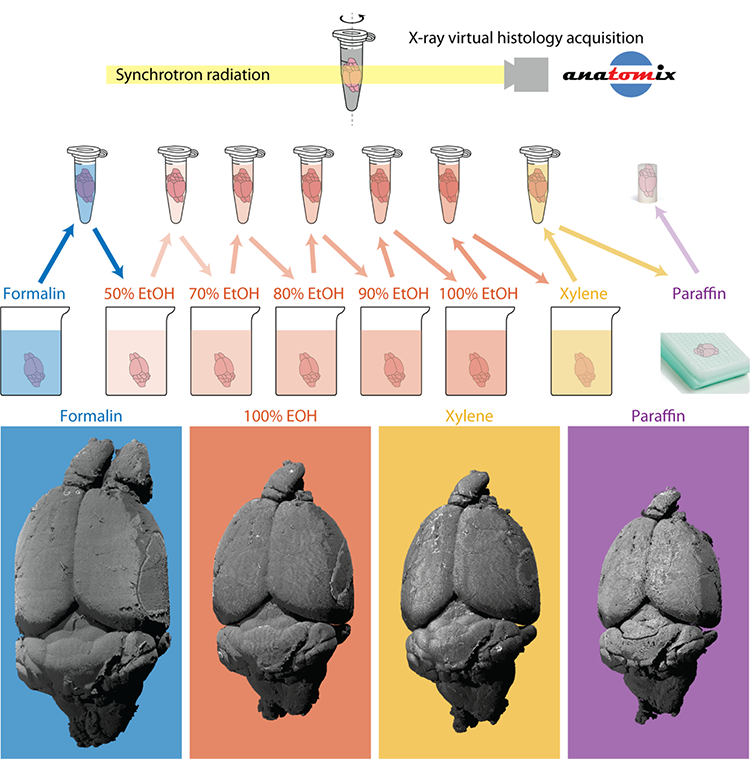
An interdisciplinary Swiss and French team of researchers from the Biomaterials Science Center at the University of Basel and University Hospital Basel, the Interface Group at the Institute of Physiology of the University of Zurich, the Swiss National Center of Competence in Research “Kidney.CH” and Synchrotron SOLEIL have now published a study in which they imaged a complete mouse brain throughout the standard histopathological preparation protocol with 3.1 µm pixel size at SOLEIL’s tomography beamline ANATOMIX. Through analysis of the 3D datasets, they quantified the composition and shrinkage of the brain. First, anatomical structures were marked and the related changes in volume were measured. The shrinkage, however, was non-uniform, therefore the correspondence between every point in the different 3D datasets had to be determined – a technique called registration.
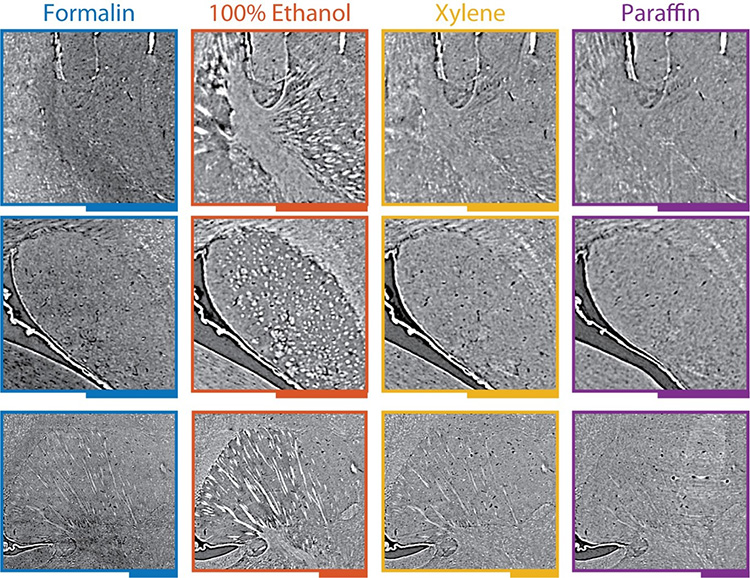
Registration allowed for the calculation of volumetric strain fields, which are 3D maps of the brain shrinkage. The scientists also transformed the datasets to match the formalin-fixed state, which effectively corrects the anatomical shape that was altered during histological preparation. Another benefit of registration is that the anatomy can be compared side-by-side to reveal surprising changes in tissue density – and thus image contrast and image quality. A remarkable change was the emergence of fiber tracts through ethanol dehydration, an effect that was reversed by subsequent xylene immersion and paraffin embedding.
Understanding and correcting the effects of tissue preparation is critical for both histology and virtual histology. The new study lays out a framework to analyze this well-known problem and allows researchers to get more out of their virtual histology datasets. For example, knowing deformations due to the embedding protocol allows researchers to make a more accurate link from high-resolution imaging in the post mortem state towards the in vivo state. Additionally, analyzing tissue density changes allows for selecting a medium to enhance X-ray contrast and bring to light anatomical features of interest. The present study was based on a mouse brain, but the methodology works on other soft tissues such as the kidney, as well as hard tissues such as tooth or bone, which are typically decalcified as part of histopathological analysis.
.