How deep-sea microbes make a life on ethane? Answers in a yellow crystal
Various hydrocarbons are naturally released in the deep-sea floor, but something in the marine sediments consumes them. The responsible are specific microorganisms, able to transform these hydrocarbons into carbon dioxide in the absence of oxygen. One of these microbial communities was recently retrieved from deep sea sediment and was shown to only use ethane and no other alkane to derive their whole cellular energy. The enzyme responsible for the ethane capture was purified from the microbial enrichment and structurally characterized by X-ray crystallography. This structure, initially solved from diffraction data collected on the PROXIMA-1 beamline, is the first of its kind and unravels the specific structural features that distinguish it from the other enzymes able to capture methane.
Geothermal activity in deep-sea sediments releases various alkanes, from the small methane to long-chain hydrocarbons. Most of these compounds are biologically consumed in the upper layers of anoxic sediments, preventing their emission in the water column and our atmosphere. Indeed, archaea living in symbiosis with bacterial sulfate reducers were showed to fully convert alkanes in CO2. From a deep-sea sediment sample collected at a 2,000 meter depth, an ethane-consuming enrichment was recently cultivated at 50 °C by Cedric Hahn and Dr. Gunter Wegener in the Max Planck Institute for Marine Microbiology (Bremen, Germany). In this culture, ethane degradation is performed by an anaerobic archaeon, Candidatus Ethanoperedens thermophilum, which appears to be unable to use any alkane other than ethane. This organism was suspected of using an ethane-fixing protein similar to the one used by methane-consuming archaea. How could this enzyme be specialized into ethane-fixation was a mystery and the best way to solve this riddle was to obtain its structure.
Different ways exist to get a snapshot of a protein structure and this type of enzymes are particularly well suited for X-ray crystallography, the specialty of Drs. Olivier Lemaire and Tristan Wagner from the Microbial Metabolism Research group (MPI, Bremen, Germany). Crystallography requires 3D crystals of the protein, whom obtention can be very laborious. Since the enzyme is known to contain an exotic cofactor (called F430) and different post-translational modifications (that are installed after the enzyme synthesis), it was imperative to purify it from the culture enrichment and not from another expression system. This is a challenge when considering the low amount of the precious culture and the fact that Candidatus Ethanoperedens thermophilum was not the only living organism there.
Because of its crucial function, the enzyme fixing ethane was very abundant in the thermophilic enrichment. It was separated from the other proteins and yellow crystals were obtained from this preparation. The crystals, frozen in liquid nitrogen, were firstly tested at PROXIMA-1 beamline at the SOLEIL synchrotron. X-rays generated by the synchrotron radiation were diffracted by protein crystals and diffraction data were collected by PROXIMA-1´s detector Eiger-16M. The first structure of an ethane-fixing enzyme was obtained from these diffraction data and exhibited the specific features that characterize it.
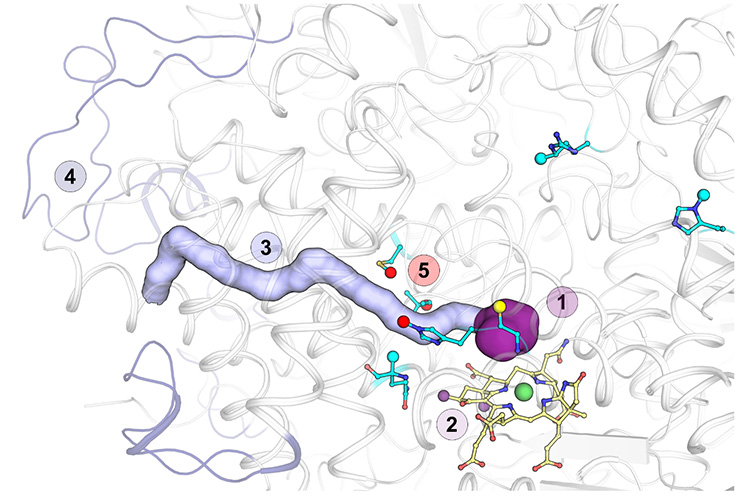
Compared to the methane-fixing enzyme, the ethane-specific protein has a wider catalytic chamber adapted to the larger alkane. The F430-cofactor, the catalyst of the reaction, contains two additional methylations, confirmed by Jörg Kahnt from the MPI for Terrestrial Microbiology (Marburg, Germany), an expert in mass spectrometry. The methylations appear to be crucial to properly position the cofactor in this widened catalytic chamber. Furthermore, the catalytic nickel atom of the F430-cofactor is coordinated by the sulfur from a methionine, in opposition to the canonical oxygen from a glutamine residue, perfectly conserved in all other enzymes of this family. This should have consequences on the cofactor properties.
The larger volume of ethane molecule compared to methane molecule prevents its efficient diffusion to the buried catalytic chamber. To overcome this issue, the enzyme harbors a 33-Å long hydrophobic tunnel bridging the outside surface to the concealed active site. This tunnel is lacking in all other characterized enzymes of this family and was confirmed by xenon-pressurization experiments performed by Dr. Sylvain Engilberge from the Paul Scherrer Institute (Villigen, Switzerland). The tunnel is stabilized by residues harboring post-translational modifications, and specific protein loop extensions. Such extensions are conserved across other ethane-fixers, allowing easier future identification of ethane-specific enzymes by their amino acid sequence.
The characterization of this enzyme unraveled several key features that makes it specific for ethane rather than other hydrocarbons. This work also opens many questions regarding the specific features of other enzymes recognizing longer alkanes. The next step is therefore to characterize these enzymes from archaea consuming longer hydrocarbons, like the butane and propane oxidizers recently described in the Max Planck Institute of Bremen.