Whooping cough: a "disordered" bait catches calmodulin and triggers the CyaA toxin
The CyaA protein is a toxin produced by the bacteria that causes whooping cough. During intoxication of the target cells, CyaA binds to calmodulin, which induces the folding of a destructured region of the toxin and its activation. This mechanism is essential for activating the toxin when it reaches its target cells - which contain calmodulin - while preventing its toxicity in the bacteria that produce it - and that do not contain calmodulin. This study, carried out by teams from the Institut Pasteur and I2BC, required the use of various biophysical techniques, including scattering of small angle X-rays (SAXS) on the SWING beamline and circular dichroism on the DISCO beamline.
Calmodulin is a highly conserved eukaryotic protein that interacts with a wide variety of proteins and enzymes. It controls their activities in response to changes in intracellular calcium concentrations. Adenylate cyclase toxin (CyaA) is one of the major virulence factors of Bordetella pertussis, the bacteria responsible for the whooping cough. CyaA contributes to the early stages of respiratory tract colonization by this bacterium in infected people. The CyaA toxin is synthesized and secreted by the B. pertussis bacteria in an inactive form. After invasion of the target eukaryotic cells, the catalytic domain of CyaA is activated by calmodulin to synthesize large amounts of cAMP that alter the physiology of the target cell. Activation of CyaA by calmodulin has been known for a long time but the molecular activation mechanism remained so far an enigma.
The researchers used an integrative structural biology approach combining several biophysical techniques to characterize structural changes in the catalytic domain of CyaA and calmodulin during their interaction. By coupling small angle X-ray scattering (SAXS), hydrogen deuterium exchange mass spectrometry (HDX-MS) and circular dichroism with synchrotron radiation (SRCD), this study has helped to clarify the relationship between structure and activation of CyaA.
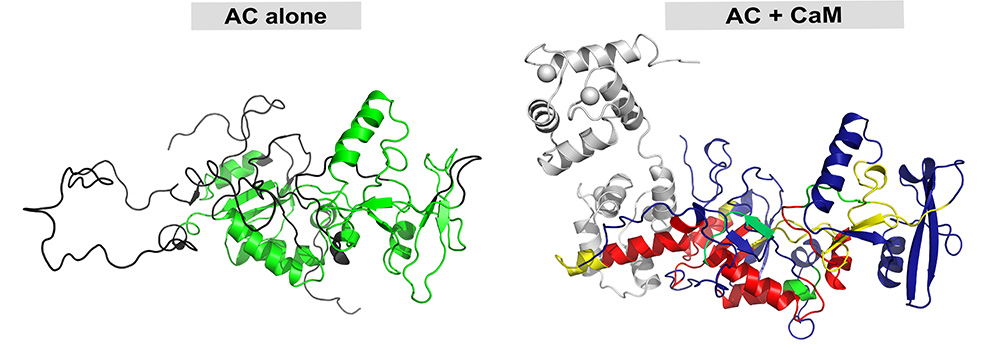
It has been found that a disordered stretch of 75 amino acids in the CyaA catalytic domain serves as bait for calmodulin capture. Binding induces significant folding within this region, a pre-requisite for CyaA activation. Beyond the region of interaction between calmodulin and the catalytic domain, the formation of the complex induces allosteric modifications and stabilizes the catalytic site distant from the interaction site. However, a catalytic loop is maintained in a very flexible state, which is essential for efficient enzyme catalysis by allowing fast binding/release of substrates (ATP) and products (cAMP and pyrophosphate). This reduction in structural disorder may be relevant to other calmodulin-activated enzymes.