SCIENTISTS SHED NEW LIGHT ON ENZYME CRUCIAL TO LIFE PROCESSES
Researchers at the University of Liverpool have revealed the crystal structure of a bacterial enzyme that offers clues on how electrons in the body move from one protein molecule to another. Their results, obtained thanks to X-ray diffraction experiments carried out, notably, on the PROXIMA1 beamline, are published in Nature.
The movement of electrons is called electron transfer and is essential for all living organisms, as it underpins processes such as respiration, photosynthesis, and detoxification.
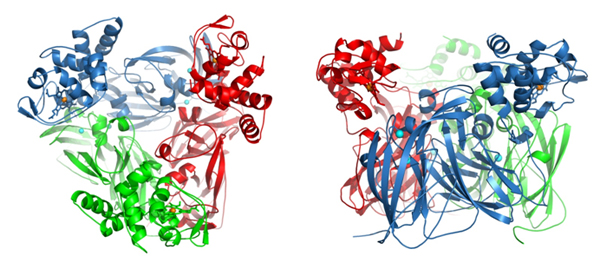
Scientists at Liverpool have examined the structure of a natural complex of the electron donor domain and the enzyme, called nitrite reductase, core. The research sheds new light on the role it plays in electron transfer, as well as the production of nitric oxide, a direct precursor to nitrous oxide, which is an ozone-depleting and greenhouse gas, 300 times more potent than carbon dioxide.
Dr Svetlana Antonyuk, from the University’s Institute of Integrative Biology, explains: “The transfer of electrons between partner proteins is key to the functioning of all living things, but our understanding of how this process occurs is limited.
“The protein complexes are very transient in nature so it is not easily possible to obtain their crystals and use the powerful method of crystallography to image them. Our system offered a major advantage as it had both the donor and acceptor proteins fused together naturally.”
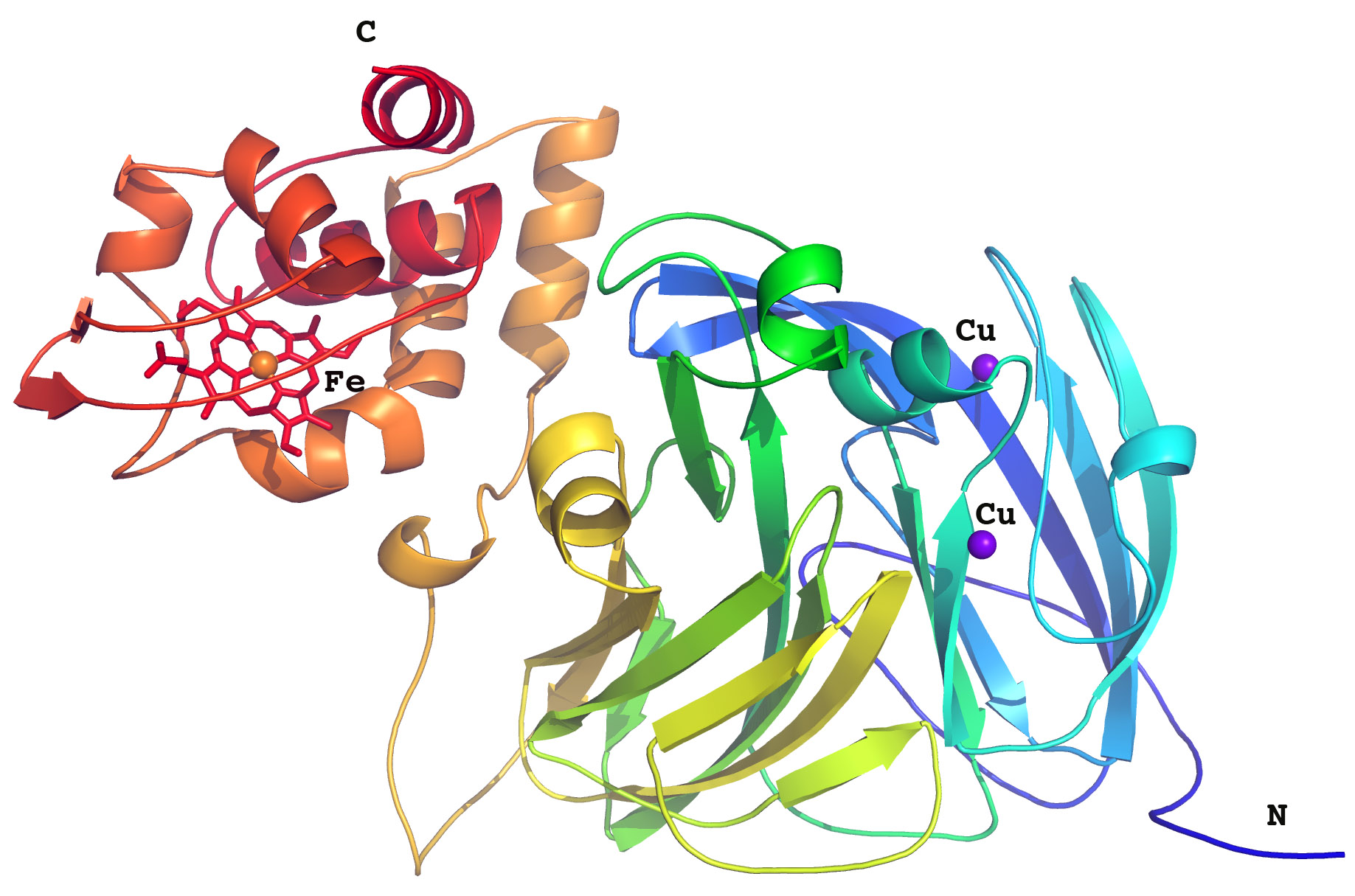
Dr Antonyuk and her colleagues worked on a bacterial nitrite reductase, RpNiR, which is very interesting because it combines, in a single enzyme, two redox partners: a cytochrome domain & a reductase domain.
Dr Antonyuk: “A striking feature of the structure is its extensive water network. Several studies have suggested the importance of water molecules in mediating transfer of electrons, but our research demonstrates it.
“In order to carry out this research we used some of the most intense X-ray synchrotron radiation sources including Diamond and SOLEIL synchrotrons [At PROXIMA1, the authors obtained data for native RpNiR in two crystal forms]. The provision of these state-of-the art facilities was critical in obtaining the high resolution of the structures of this tethered complex and allowed us to see every atom in the molecule.”
Prof. Samar Hasnain, co-author of the Nature publication, adds: “The openness of SOLEIL to international users from its inception has been very helpful to many of us in addressing challenging problems in structural biology".
Dr Antonyuk concludes: “Our findings will provide important fundamental insight into life processes.”
Access to SOLEIL beamlines (for excellent European projects in the life sciences) is facilitated by the BioSTRUCT-X FP7 project of the European Commission. The deadline for the next call for projects is 30th April.
The research is funded by the Biotechnology and Biological Science Research Council (BBSRC).