Regulation of gene expression - Take off your cap!
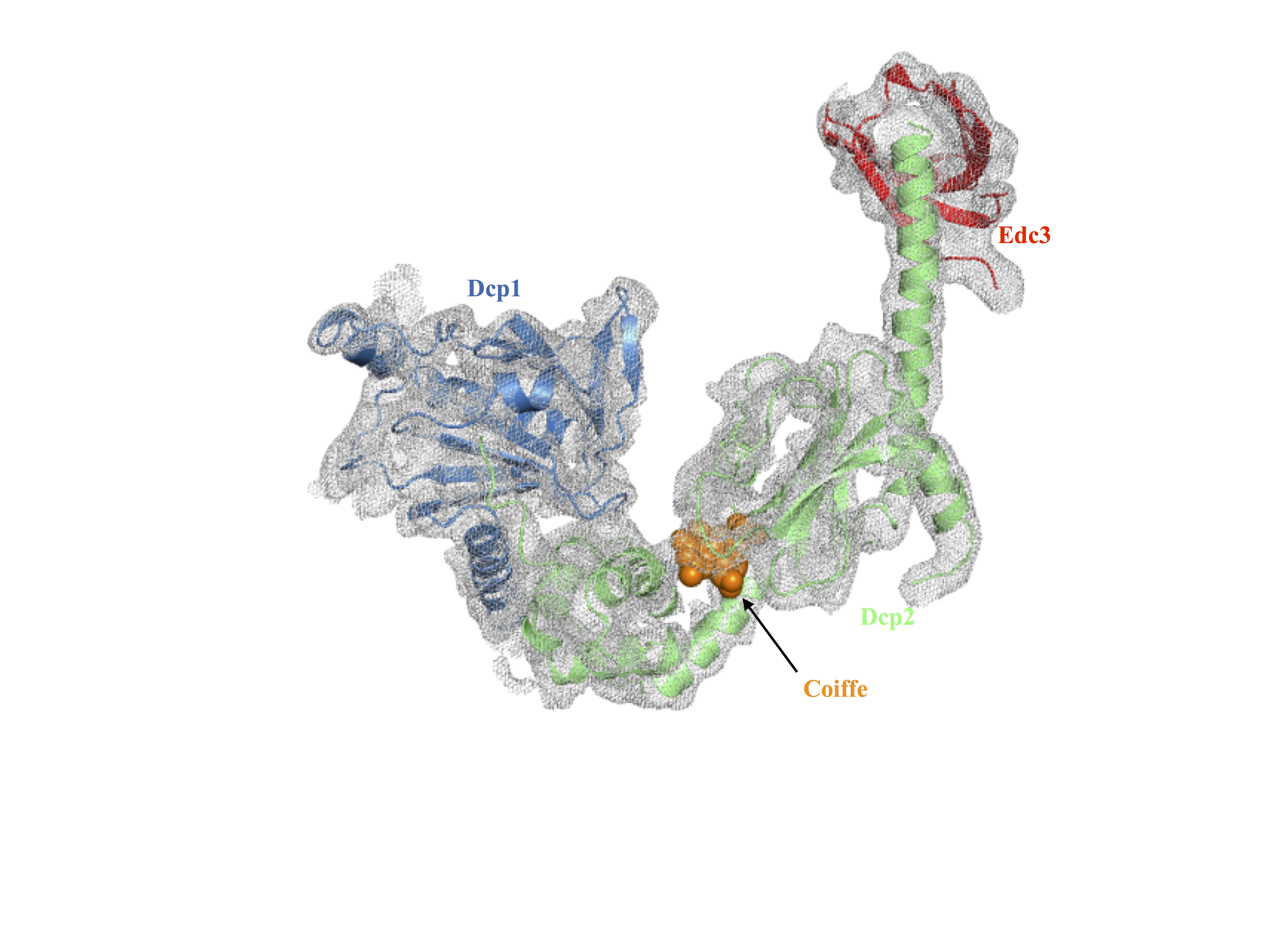
Through use of the PROXIMA-1 and PROXIMA-2A beamlines, researchers of the Ecole Polytechnique (CNRS, Palaiseau) and IGBMC (CNRS, INSERM and Strasbourg University) have determined the structure of a complex of proteins: the enzyme Dcp2, associated with its two 'helper molecules', the cofactors Dcp1 and Edc3. The structure of this complex has revealed the mechanism of action of Dcp2, implicated in the removal of the messenger RNA (mRNA) cap, a reaction essential for the degradation of mRNA molecules during the course of protein synthesis. Until now the molecular mechanism of Dcp2 had not been discovered.
These results are published in the journal Nature Structural and Molecular Biology.
In a cell, the production of proteins takes place in several steps. The first is the synthesis of 'messengers', i.e. mRNA molecules synthesised by copying a given gene. These mRNA molecules are next 'translated' into proteins following the rules of the genetic code (i.e. a given amino acid of the protein is produced corresponding to each mRNA nucleotide triplet).
In eukaryotes, the mRNA molecules are protected against uncontrolled elimination by stabilising elements at each of their ends: a cap at one end (5' end) and a polymer of adenosine (or poly(A) tail), at the other (3' end). The objective of the teams of Marc Graille (Ecole Polytechnique) and Bertrand Séraphin (IGBMC) is to understand the mechanisms of degradation of the mRNA and their contribution to the regulation of the expression of genes.
The first step in the degradation pathway of mRNA is generally the shortening of its poly(A) tail, or deadenylation. This step is generally followed by the cleavage of the cap which triggers the irreversible and complete destruction of the mRNA. The cleavage of the cap also prevents any further production of proteins based on this mRNA. Indeed, mRNA must have a complete 5' end so that the translation factors, proteins which help the ribosome during the initiation of the translation of the mRNA, bind correctly to this mRNA. The cleavage of the cap is catalysed by the enzyme Dcp2. Although this factor has been known about for more than 10 years, its mechanism of action remains mysterious because this enzyme is scarcely active in vitro and requires numerous co-factors to act in vivo. Several structures of Dcp2 have been described, but they do not explain its activity as a catalyst.
The structure of Dcp2 associated with its co-activators Dcp1 and Edc3 and bound to the product of the cleavage reaction of the cap
The team led by Marc Graille has succeeded in crystallising the complex Dcp2-Dcp1-Edc3 in the presence of a cap analogue. The data collected on the PROXIMA-2A beamline using the Eiger 9M detector have been used to determine an experimental electron density map with a resolution of 4Å. This led to the accurate positioning of the different components of this complex so that its organisation was revealed. In this structure, two Dcp2 domains hold a nucleotide in a pincer grip, where the nucleotide represents the cleavage product of the mRNA cap by Dcp2, explaining the biochemical data published previously. The structure also shows that there is an allosteric regulation mechanism of Dcp2 by its activator Edc3: Edc3 induces a change in the conformation of Dcp2, which favours its activity and the recruitment of mRNA.
These results offer new perspectives in the understanding of the cap cleavage mechanism and may allow development of the inhibitors of the enzyme which play a key role in the degradation of mRNA, essential for the survival of eukaryotic cells.