A new tool in the fight against multi-resistant bacteria
Scientists from France, Germany and Italy, in collaboration with the PROXIMA-2A beamline, have designed a new class of active molecules that inhibit one of the major mechanisms involved in the phenomenon of multidrug-resistant bacteria (MDR). These results could represent a new strategy in the fight against the scourge of MDR.
Before the antibiotic era, a small cut on the finger could lead to a fatal infection. In 1928 Sir Alexander Fleming accidentally discovered penicillin - for which he was awarded the Nobel Prize - and since then this miracle drug has saved millions of lives. However, the overuse and misuse of antibiotics in agriculture and aquaculture, as well as their overprescription to the public, have endangered their efficacy, contributing to the development and dissemination of Multi-Drug-Resistant (MDR) bacteria. The World Health Organization warns that drug resistant infections could cause 10 million deaths each year by 2050.
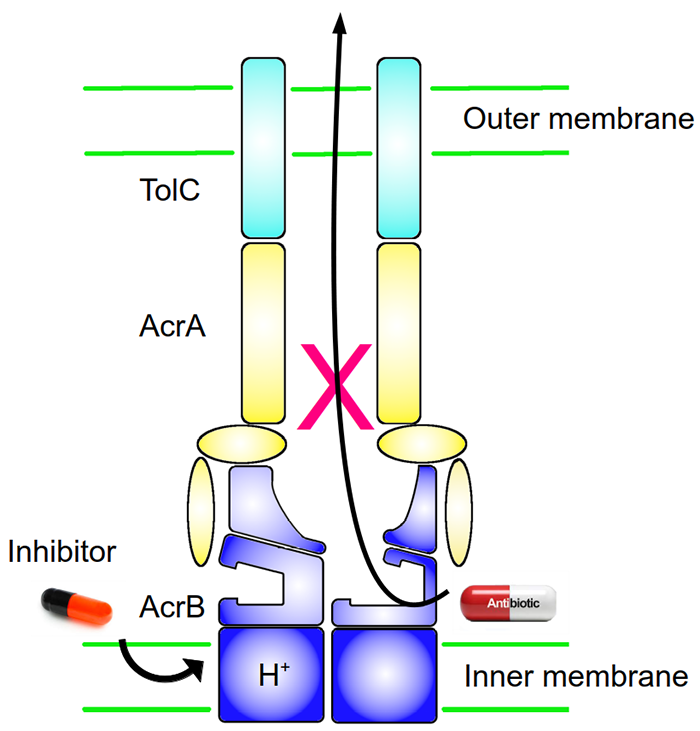
One of the major strategies that bacteria employ to cope with the presence of antibiotics is the MDR “efflux pump”. Formed by proteins located in the membrane(s) on the surface of the bacteria, this pump will, like a kind of vacuum cleaner, evacuate the toxic substances present in the bacteria, including antibiotics. However, "aspiration" - that is, the mechanism of transport of antibiotics out of the bacteria - is a complex process. Many studies have shown that the "AcrAB-TolC" efflux pump of the Escherichia coli bacterium - which causes, among other things, turista, or traveler's diarrhoea - uses several different "tubes" (transport pathways) to suck up the antibiotics present in the bacterium, and then transfers them to its "dust bag": a binding pocket, so called because the antibiotics, notably, link to it. Finally, this dust bag is emptied: the antibiotics are expelled.
For years, research aimed at inhibiting efflux pumps has mainly focused on this binding pocket, the objective being to find molecules that can bind to this pocket instead of the antibiotics, thus preventing their expulsion. Scientists have thus designed several agents that block the "dust bag" (inhibitors) in order to prevent aspiration. However, all these inhibitors have failed to reach the market due to their lack of potency or, more notably, their toxicity to the patient.
Recently, scientists from the Pasteur Institute of Lille, the Goethe University of Frankfurt and the University of Cagliari have joined forces to design inhibitors that work by cutting off the energy supply needed for antibiotic "aspiration" - this energy is provided by a proton motive force present in the bacterial membrane. Initially, 1280 compounds were screened for the inhibition of AcrB, which is a model system for studying efflux pumps. Only one compound was identified to inhibit the AcrB activity, however it lacks potency. Subsequently, by analysing its structure-activity relationships, the scientists synthesized 11 other compounds. To understand how they work, the scientists mixed each of them with the AcrB protein. They then determined the 3D structure of the pump/inhibitor complex formed, using X-ray crystallography methods. These experiments were carried out on the PROXIMA-2A beamline at SOLEIL.
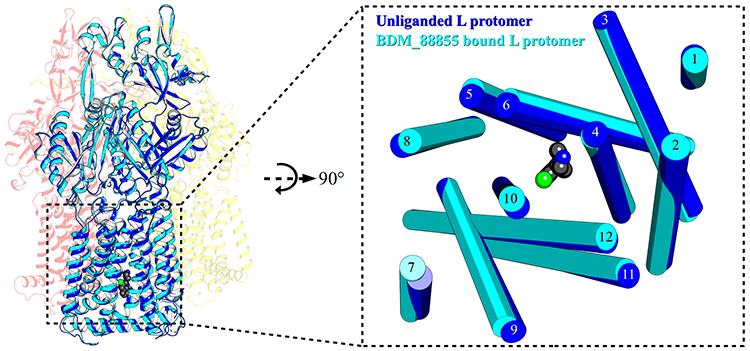
Left: 3D structure of the AcrB protein bound to an inhibitor molecule (bottom center, black and green balls), attached in the binding pocket that prevents protonation.
Right: zoom on the inhibitor in the center of the pocket.
The analysis of the data obtained allows the visualisation, on the atomic scale, of where and how the inhibitors attach to the pump and causes the "power cut" during the aspiration of the antibiotics. The scientists thus demonstrated that the inhibitors bind to a previously unknown pocket of the AcrB pump, located in the transmembrane part of the bacteria, close to the “proton relay site” that supplies the pump with energy. These results suggest that inhibitors could work by preventing either a conformational change in the pump - necessary for its proper functioning - or impeding the proton relay, which would lead to a malfunction of the antibiotic expulsion mechanism.
These discoveries constitute a new strategy in the fight against multi-drug resistant bacteria.