Microchips to explore the 3D architecture of biomolecules
Crystallography is one of the most widely used methods for the study of the 3D architecture of biomolecules (proteins, RNA, DNA). However, the most delicate stage in this technique is the production of crystals from the biomolecule from which the 3D image is to be determined.
A team of French researchers, in collaboration with European colleagues and 4 beamlines, including PROXIMA 2A at SOLEIL, has designed a new microfluidic chip that miniaturizes and optimizes the process of identification of crystallization conditions. It also enables the direct analysis of crystals by serial crystallography using the latest innovations in synchrotron radiation.
Their results were published in May 2019 in the IUCrJ journal.
Crystallography plays a central role in contemporary biology because it enables the visualization of the 3D architecture of biomolecules - proteins and nucleic acids - and provides insights into the understanding of their cellular functions at the atomic scale. This technique consists in growing crystals composed of the biomolecule in question, followed by an analysis of the diffraction properties of these crystals under X-ray, most often using synchrotron radiation, to determine the spatial arrangement of the atoms that compose this biomolecule. Despite advances in crystallography in recent decades, obtaining quality crystals from a limited amount of biomolecules remains a bottleneck in this type of study. This process generally involves sampling of chemical and physical space by screening hundreds of cocktails composed of buffers at different pH levels, various crystallants - salts, alcohols and polymers - at different temperatures to identify a favourable condition for crystal growth.
Following the introduction of the first microfluidic systems dedicated to high throughput (HTP) screening 15 years ago, the sample volume required was reduced to a few (tens of) nanolitres per test. Consequently, microfluidics was immediately recognised as a major breakthrough, especially for biochemists working with samples difficult to purify in large quantities, such as large biological complexes or membrane proteins. However, despite their potential, these microfluidic technologies struggle to be adopted by the global community for crystal growth. This is partly explained by the cost of these microsystems and their associated equipment, but also by the difficulty of extracting fragile crystals from chips, or the requirement to reproduce them using conventional crystallization methods before they can be subjected to crystallographic analysis.
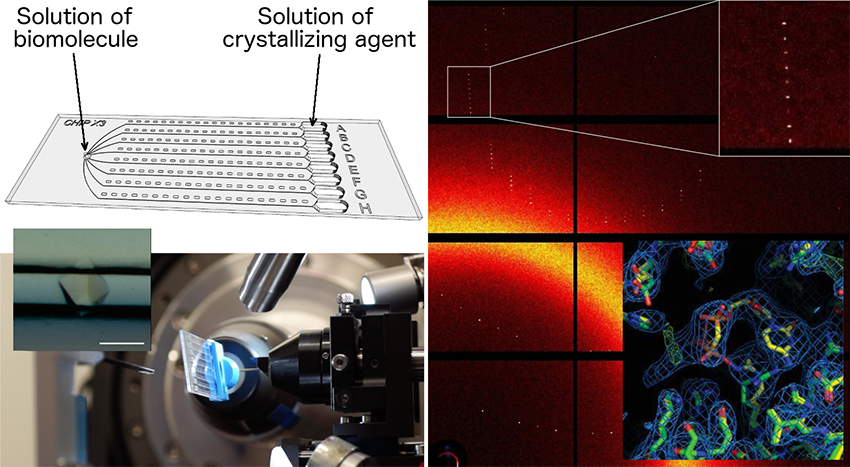
In order to expand the interest and functionality of microfluidic chips beyond crystallization and HTP screening, several teams have investigated the possibility of analysing the crystals directly in their microfluidic environment. In a recent article, a French team (CNRS – the University of Strasbourg), in collaboration with teams from Grenoble, Marseille, Leipzig (DE), Granada (ES) and three European synchrotrons (SOLEIL, ESRF, SLS), have developed a versatile and cost-effective microfluidic chip for the production and characterization of crystals. This chip was initially designed to miniaturize and facilitate the identification of crystal growth conditions using the counter-diffusion method and its efficient self-optimization process.
The latest version of the chip, called ChipX3, incorporates several improvements in terms of sample injection, reservoir loading and adaptation to X-ray analysis. With ChipX3, it is effectively possible to:
1) produce crystals including by seeding,
2) soak them in situ for the introduction of ligands,
3) visualize them by fluorescence imaging,
4) determine the 3D structure of the biomolecule they are composed of by on-chip serial crystallography at room temperature.
A genuine biocrystallographic Swiss army knife!