How coronavirus HKU1 forces its way into cells
Scientists from the Pasteur Institute have pinpointed the precise binding sites for the coronavirus HKU1 in respiratory cells, providing a detailed picture of the infection mechanism used by the virus. Their findings shed light on other animal species that may host the virus and pave the way for the development of infection-blocking antibodies.
SOLEIL's PROXIMA-1 and PROXIMA-2A beamlines contributed to these results.
The contribution of SOLEIL
On SOLEIL's PROXIMA-1 and PROXIMA-2A beamlines, Institut Pasteur scientists analyzed the crystals of complexes containing the receptor molecule that enables HKU1 to bind to cells, called TMPRSS2 (only the part recognized by the virus), the part of the virus’ spicule that binds to this receptor protein, and antibodies (nanobodies).
From the various X-ray diffraction data sets collected, three crystallographic structures were obtained - accessible in the ProteinDataBank – thanks to data collected on PROXIMA-1 beamline, which helped to specify, on an atomic scale, the site recognized by the HKU1 spicule and the epitopes (interaction regions) of the cellular receptor to which the antibodies bind. These are the 3 structures presented in the Cell journal publication on this work.
Find out more about these results from Institut Pasteur researchers, published in the Journal Cell, here
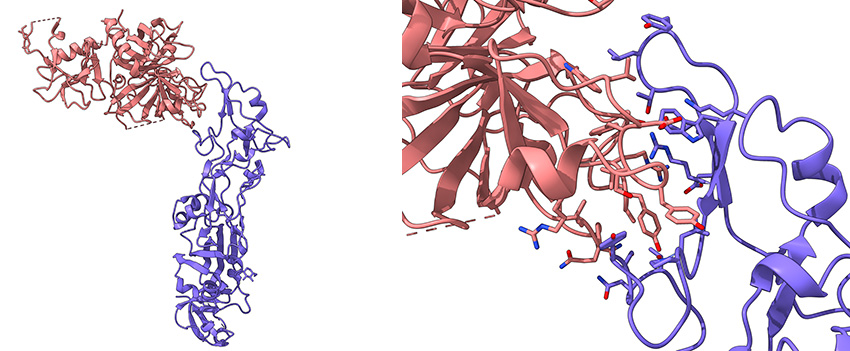
Left: Schematic representation of the complex between part of the TMPRSS2 cell receptor (pink) and the receptor-binding domain of the HKU1 coronavirus spike protein (purple).
Right: Zoom on the interaction region of these two proteins.