From Archaea membranes to industry: the key to the mevalonate biosynthesis reaction revealed
Researchers from the Max Planck Institute for Terrestrial Microbiology (Marburg, Germany), the Institute for Structural Biology (Grenoble, France) and the École Normale Supérieure (Lyon, France) have determined the structure of the enzyme complex responsible for the synthesis of mevalonate, the essential building block constituting Archaea membranes. Moreover, they have discovered the "molecular trick", which allows this reaction to occur despite a very unfavourable first step in terms of energy. This discovery is also of interest to the industrial sector, as mevalonate is a molecule used in the synthesis of numerous chemicals.
The lipid membranes of the archaea are composed of isoprenes produced from mevalonate. Three enzymes are involved in the biosynthesis of mevalonate: thiolase, HMG-CoA synthase (HMGCS) and HMG-CoA reductase. The first reaction of this synthesis catalyzed by thiolase is highly endergonic (i.e. the reaction requires a supply of energy and thus the flow of production is very low), which is in contradiction with the rapid growth of the archaea, and consequently their lipid synthesis.
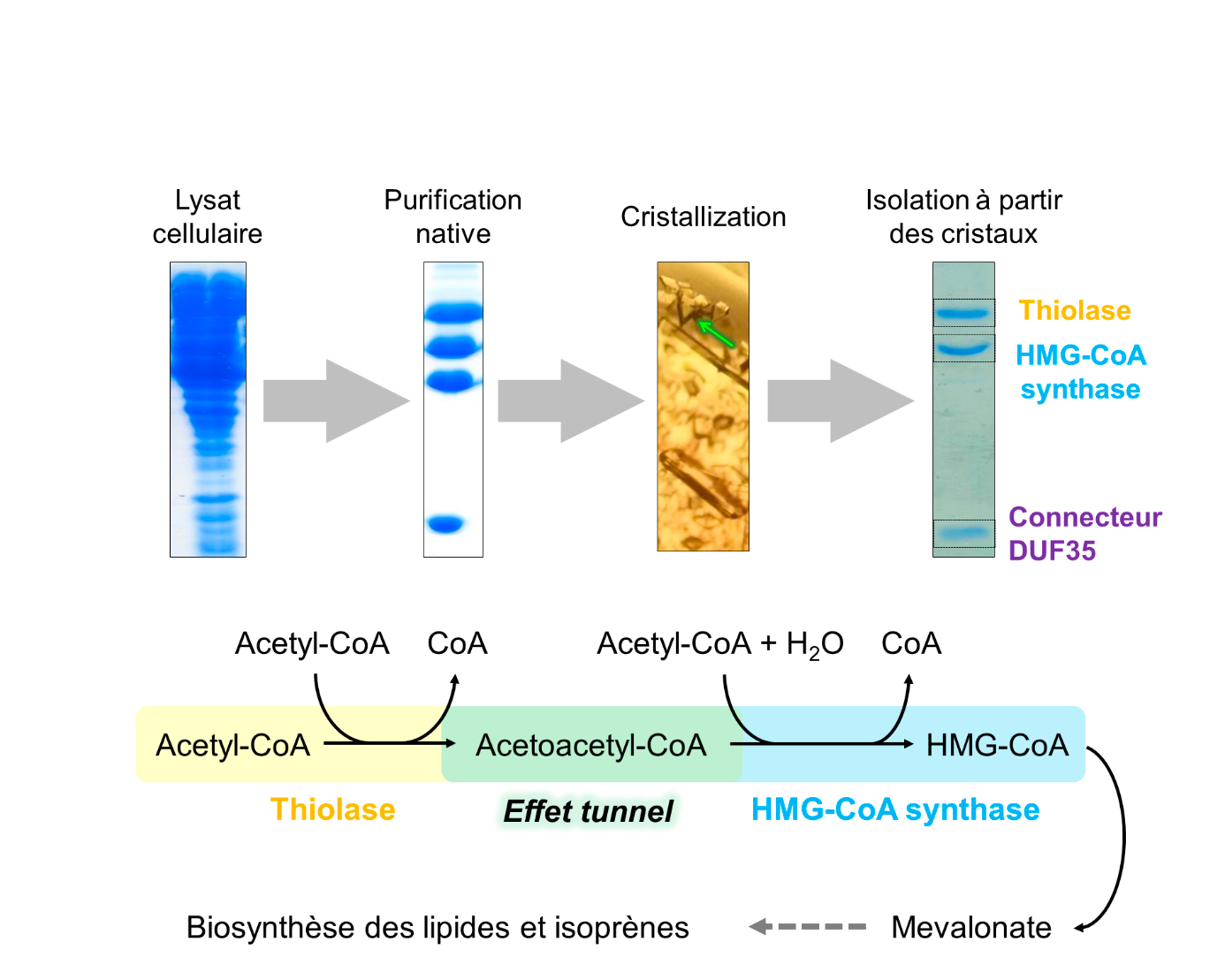
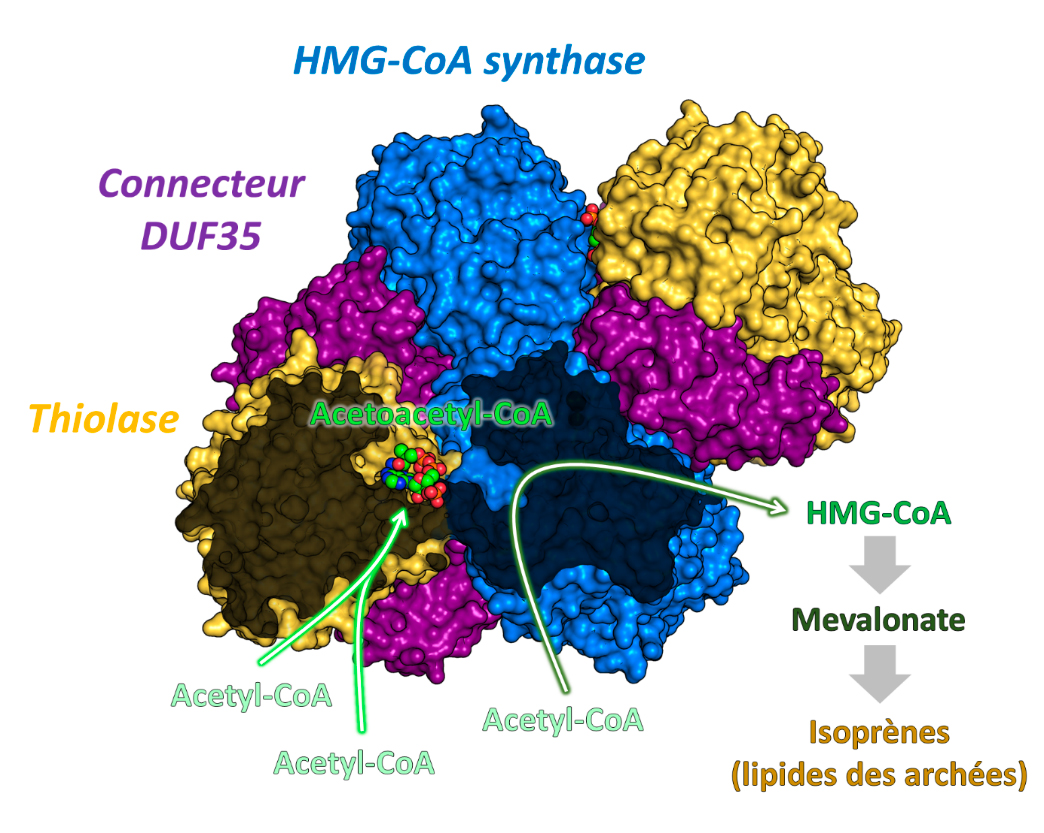
Researchers from Seigo Shima’s and Tobias Erb's team[s?] at the Max Planck Institute for Terrestrial Microbiology in Marburg, Germany, in collaboration with researchers from Eric Girard's team at the Institute of Structural Biology, Grenoble, and Olivier Maury at the École Normale Supérieure, Lyon, have joined forces to unveil the molecular trick that allows archaea to accelerate the speed of the thiolase. By purifying the thiolase directly from archaeal cells, the researchers found that the enzyme formed a natural complex with HMGCS, the second enzyme in the biosynthesis of mevalonate (Figure 1). Both enzymes are held together by a third smaller connecting protein (called DUF35). Using Tb-Xo4, a new tool that induces crystallization and greatly facilitates the structural determination via the presence of a lanthanide atom, coupled to the X-ray diffraction screening capabilities on the protein crystallography beamline PROXIMA-2A, the structure of the thiolase/HMGCS/DUF35 protein complex was resolved (Figure 2).
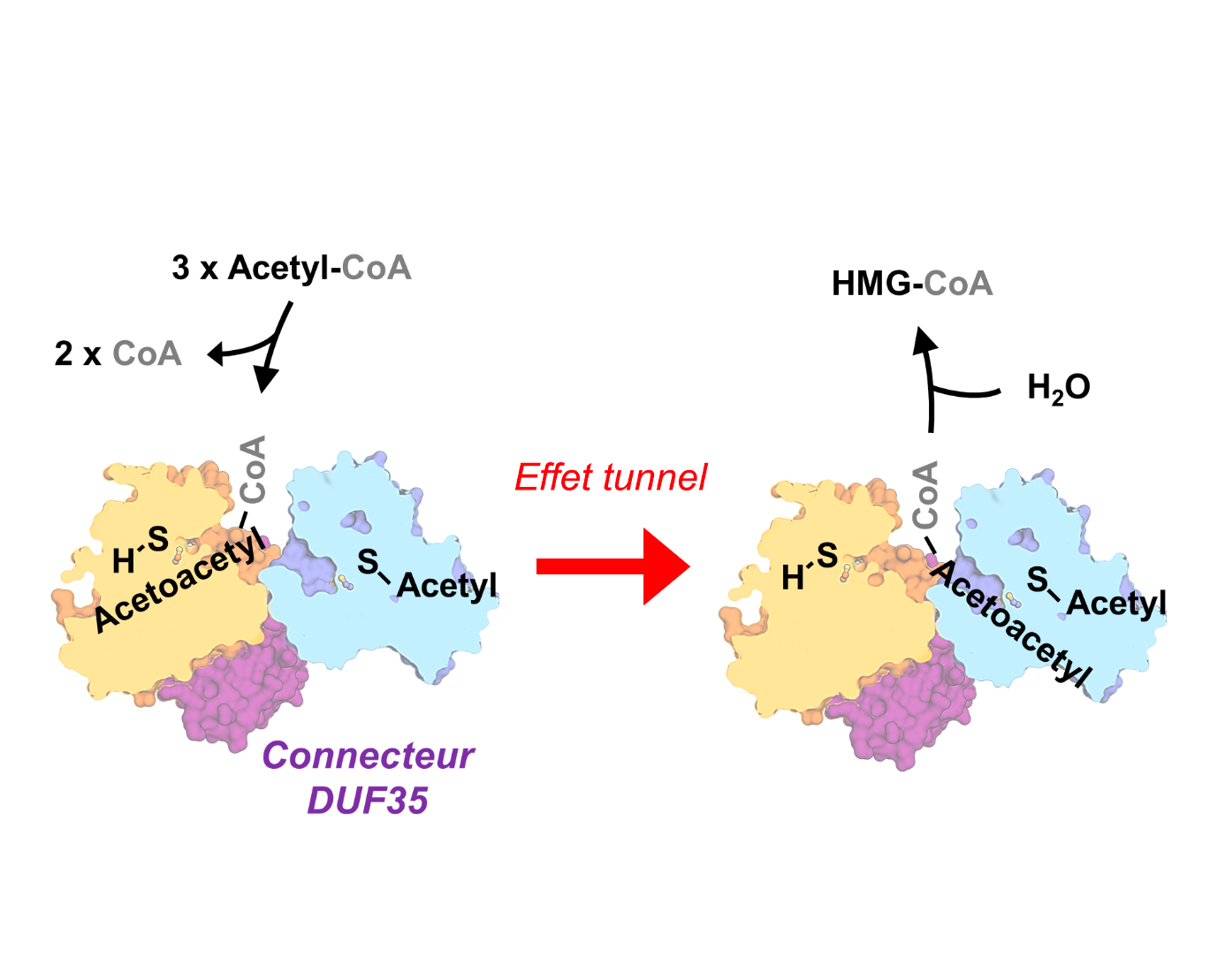
The structural and biochemical study revealed the key to the reaction: thiolase and HMGCS share the same substrate-binding site for Acetyl-CoA. This unique site enables a "tunnel effect", which shifts the reaction favourably and consequently increases considerably the synthesis speed. In other words, the endergonic thiolase reaction is pulled by the exergonic (energy releasing) HMGCS reaction (Figure 3).
This discovery is of prime importance for many industries: mevalonate is a precursor molecule that allows the synthesis of many chemical substances, such as synthetic rubber.