Antivirals alter the elastic properties of the hepatitis B virus capsid
One strategy for curbing the proliferation of pathogenic viruses is to disrupt their assembly as they multiply in infected cells, in particular with small molecules known as assembly modulators. Modulators have been developed to target and deform the capsid of the hepatitis B virus, making it no longer viable for virus propagation. However, the physical mechanisms of action of these antivirals remain poorly understood.
An international consortium has shown that the capsid deformations caused by these modulators can be explained by an alteration in the elastic properties of the capsid components. Time-resolved X-ray scattering on the SWING beamline has made it possible to measure the interaction energies induced by modulators between capsid proteins.
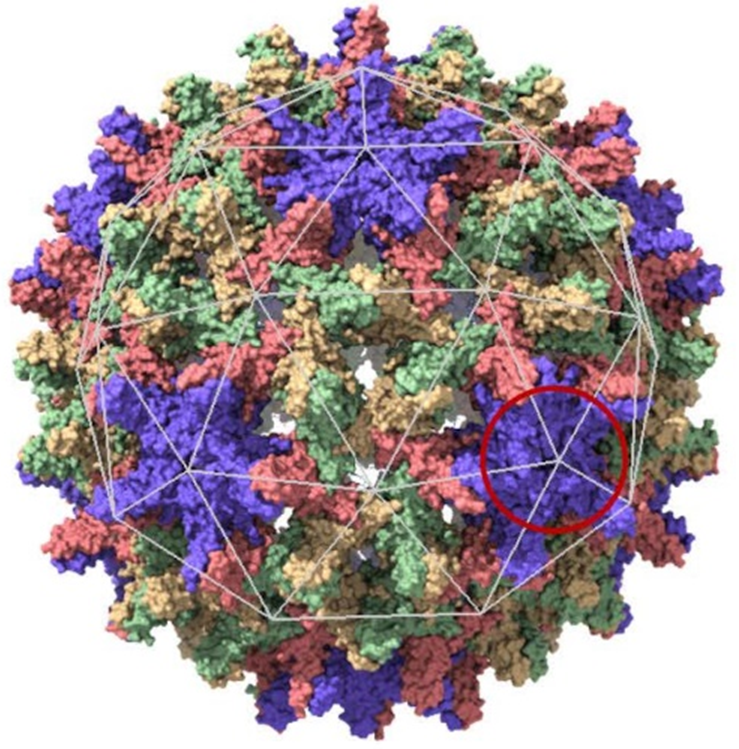
Over 250 million people are chronically infected with the hepatitis B virus (HBV), mainly in Africa and South Asia, due to the lack of systematic vaccination. HBV is the cause of severe pathologies such as liver cirrhosis and hepatocellular carcinoma, leading to the death of almost a million patients every year. HBV is an enveloped virus comprising a protein shell - the capsid - arranged in an icosahedral symmetry structure (20-sided structure, Figure 1), protecting the genome in DNA form. During a virus life cycle, the subunits - a subunit being a dimer of the capsid protein - synthesized by the machinery of the infected cell self-assemble into capsids before crossing the plasma membrane to infect neighboring cells.
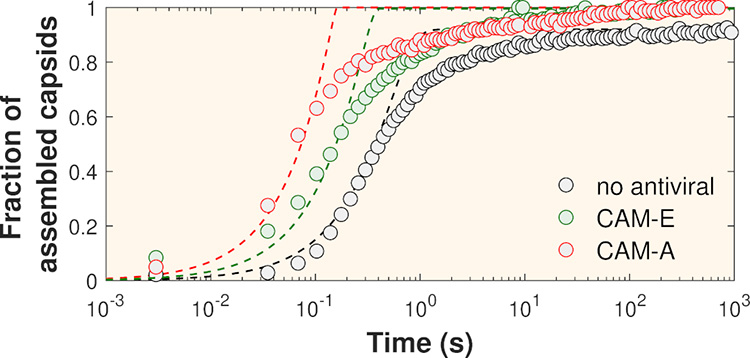
Researchers at the Université Paris-Saclay have reproduced in vitro the self-assembly process of the HBV capsid in the presence of two types of assembly modulators, CAM-E and CAM-A (CAM for Capsid Assembly Modulator). When purified viral capsid proteins, initially stored in a low ionic strength solution, are suddenly placed in a solution with a physiological ionic strength of around 150 mM, they spontaneously assemble, resulting in a mixture of capsid and unassembled proteins. Using time-resolved small-angle X-ray scattering (TR-SAXS) on the SWING beamline, the researchers followed the self-assembly kinetics with high spatio-temporal resolution. Figure 2 shows the mass fractions of assembled capsids as a function of time deduced from TR-SAXS measurements. Using a phase transition kinetic model, the interaction energies between subunits were found to increase from 9kBT without antiviral agents - kBT being the thermal energy - to 18kBT in the presence of CAM-A, meaning that the interaction between capsid-forming proteins becomes twice as strong in the presence of this antiviral agent.
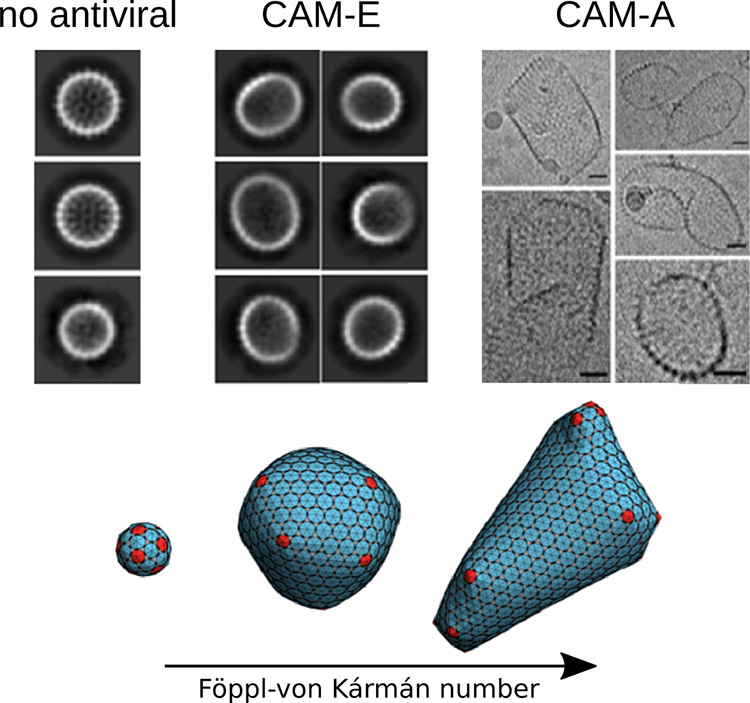
The final objects were imaged by cryotransmission electron microscopy (cryoTEM) and found to be either slightly ellipsoidal in the presence of CAM-E, or very strongly deformed and larger in size with CAM-A than in the absence of antivirals (Figure 3).
Finally, a team from the University of California at Riverside simulated capsid assembly using a set of triangles (see Figure 1) and varying the ratio between the stretching energy and the bending energy of capsids (Föppl-von Kármán number). In this way, they obtained morphologies very similar to the capsids imaged by cryoTEM (Figure 3).
This study reveals that modulators considerably enhance the attractive interactions between HBV capsid subunits, most certainly by increasing the hydrophobic contact surface. Evidence not presented here further suggests that modulators like CAM-A lower capsid curvature energy, probably due to their steric hindrance which confers flexibility in the angle formed by adjacent subunits. In addition to providing a better understanding of antiviral mechanisms of action, these results could open up prospects for the design of virus-derived nanocapsules with controllable morphologies.