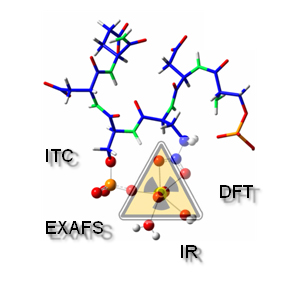
Which protein is responsible for uranium accumulation in the skeleton?
Radiochemists from CNRS/Université Paris-Sud Orsay (IPNO) and CNRS/ Université de Nice-Sophia Antipolis (ICN) and biologists from the life science CEA division (DSV) collaborated with the MARS beamline and were able to address this fundamental question.
The large development of nuclear industry for civil and military applications during last century, has increased the risks of hazards due to radionuclide release or exposure. In this context, toxicological studies on the impact of radionuclides, and in particular uranium, are of crucial importance.
Concerning uranium toxicity, medical studies have shown that most patients exposed to uranium have developed kidney failure or bone cancer. In this latter case, it is conjectured that at a molecular level, Osteopontin (OPN), a newly identified human target protein, might play a big role in uranium in vivo accumulation in bone growth regions which post-facto can induce bone cancer. The description of OPN's interaction with uranium both structurally and thermodynamically is thus of great importance to get more insights on uranium bone accumulation and for developing possible remediation methods to uranium bone contamination.
Coupling experimental techniques and theoretical calculations
Studies carried on the OPN's amino-acid sequence have identified an active site (codenamed H8V) which plays a major role in bone matrix growth inhibition. Since proteins are complex by nature, the investigation consisted in studying the interaction of uranium ions with an 8 amino acids molecule (octapeptide) mimicking the identified active site.
The experimental approach we used, coupled spectroscopy, in particular Extended X-Ray Absorption Fine Structure spectroscopy (EXAFS) with synchrotron radiation, thermodynamics and theoretical calculation (DFT).
Initially thermodynamical aspects and speciation of the uranium interaction with the peptide molecule were investigated using both Time Resolved Laser Fluorescence Spectroscopy (TRLFS) and Isothermal Titration Calorimetry (ITC). Both techniques revealed that only one uranium atom, in oxidation state VI, is fixed onto the studied active site with quite a strong apparent chemical affinity (Log Ka = 3) which verifies our assumption that uranium interacts with OPN.
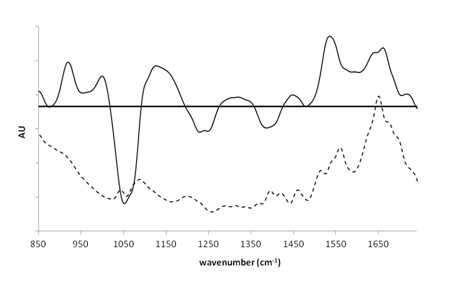
Ensuite, la spectroscopie infrarouge à transformée de Fourier en réflexion totale atténuée (ATR-FTIR) a été utilisée pour comprendre l'environnement moléculaire du complexe uranium(VI)-peptide afin de déterminer le mode de liaison du groupe carboxylate du peptide et de détecter l'interaction attendue avec le groupe phosphate de ce peptide. La présence sur le spectre différentiel (Fig1, droite) d'une bande négative à 1383 cm-1 et d'une bande positive à 1459 cm-1 est caractéristique d'un mode de liaison bidentate (2 liaisons) pour le carboxylate. D'autre part, la présence de bandes à 1246 cm-1, 1056 cm-1 et 1124 cm-1 est caractéristique d'un changement géométrique dans les groupes phosphates induit par la liaison de l'uranium.
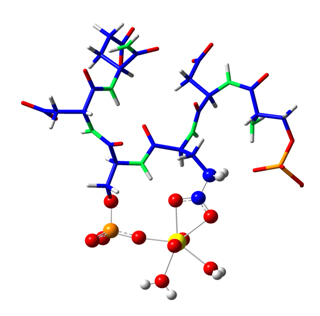
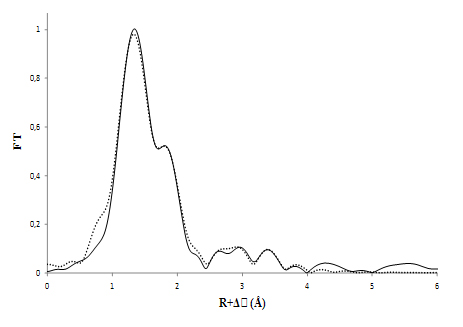
Enfin, des études structurales supplémentaires ont été effectuées pour sonder l'environnement de coordination de l'uranium en utilisant la spectroscopie EXAFS. Ces expériences ont été réalisées sur la ligne MARS, dédiée à l'étude des matériaux radioactifs. En parallèle, ont été testés divers modèles structuraux qui ont été développés en accord avec les précédentes données spectroscopiques et thermodynamiques, et optimisés par des calculs théoriques ab-initio. Il a été alors possible de faire un ajustement sur les données expérimentales EXAFS en utilisant la courbe correspondant à l'une des structures identifiées par la théorie.
How does osteopontin bind uranium
The EXAFS fitting procedure indicated that uranium was bound to a phosphoryl and a carboxyl functional group of the peptide molecule, with an average distance of 2.90 and 3.87 Å between the uranium atom and the carbon and phosphorus atoms respectively. These distances are indicative of an inner-sphere complex with one oxygen atom shared between the uranium and the phosphoryl group and two oxygen atoms from a single carboxyl group (bidentate coordination mode). The coordination sphere was completed with 2 water molecules.
The pertinence of this approach was also emphasized by EXAFS measurements done on the full protein. Indeed, the coordination environment observed with the octapeptide’s active site was strikingly similar to that formed with the full protein at the exact same conditions.
This study not only managed to provide insight into uranium bone accumulation but also widened the classical study scope found in the literature. Indeed most uranium studies up to now have dealt with previously known protein metal binding sites, thus ignoring all possible uncharacterized binding site sequences within proteins. In future works the scientists hope to identify and characterize new protein uranium targets in order to rationalize the inherent mechanisms of bioaccumulation that govern its toxicity.