Monitoring the motion of a microbiome enzyme involved in methane biosynthesis
The largest and most diverse family of enzymes are the so-called radical SAM enzymes. They catalyse a large number of major biochemical reactions often unprecedented in living cells, notably for the biosynthesis of antibiotics or vitamins. Despite their importance, their wide distribution in living organisms and their key biological roles in bacteria but also in humans, these enzymes are difficult to study as they are oxygen and light-sensitives.
Scientists from INRAE (MICALIS, ChemSyBio), the PROXIMA-1 beamline and Aix-Marseille University-CNRS, have investigated the role of Mmp10, an enzyme involved in methane biosynthesis. Methane is a major greenhouse gas but it also plays an important role as a source of bioenergy and in the microbiome*.
The production of methane by anaerobic archaea is responsible for two thirds of the world's methane emissions, a large part of which comes from marine sediments and mammalian microbiome. The final step in methane production involves the enzyme "methyl-coenzyme M reductase" (MCR), whose activity is regulated by another enzyme, called Mmp10. Mmp10 introduces a key post-translational modification of MCR active-site, which is critical for its activity and methane production. This modification -the methylation of an unactivated carbon-atom - is a particularly challenging reaction that only enzymes similar to Mmp10, a family of enzymes called "vitamin B12 -dependent radical SAM enzymes", can catalyse. However, since their discovery ten years ago, we don’t know how these enzymes catalyse such challenging modification.
Researchers from INRAE (MICALIS, ChemSyBio, Université Paris-Saclay) in collaboration with the PROXIMA-1 beamline and Aix-Marseille University-CNRS have undertaken the biochemical, spectroscopic and structural study of Mmp10 in order to decipher its 3D structure and reaction mechanism.
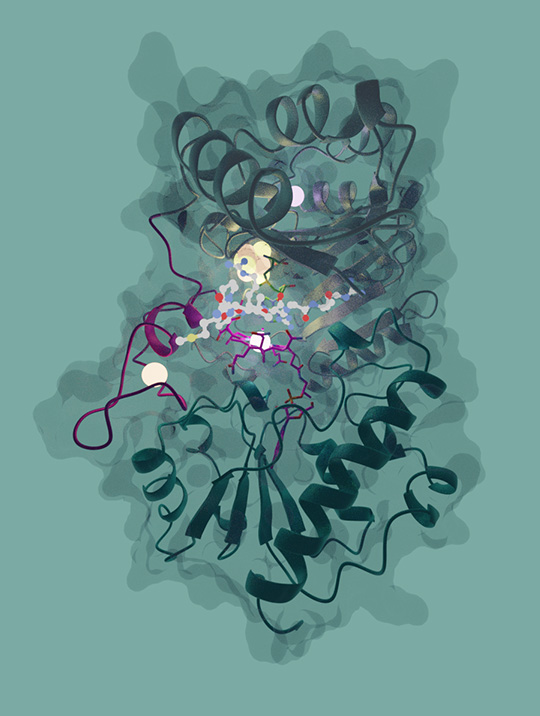
Their novel results show that Mmp10 possesses an unprecedented three-dimensional structure among known enzymes, with notably four distinct sites containing metal atoms, including two iron-sites (a 4Fe-4S cluster and a mononuclear iron), a sodium cation and a cobalt atom (Figure 1). Scientists have demonstrated that all these metallic centers are critical for enzyme activity.
In addition, for the first time, it was possible to get several "snapshots" of Mmp10 at different steps of the reaction, i.e. to obtain the 3D structures of the enzyme in the various conformations sequentially adopted through the reaction it catalyses. The scientists were hence able to capture the enzyme in motion and obtain a “movie” of the reaction. Thanks to this elegant and multidisciplinary approach, it was possible to demonstrate that the enzyme undergoes unprecedented rearrangements, mainly in its active-site, through catalysis.
This breakthrough discovery explains how Mmp10 carries out the methylation reaction of the MCR enzyme by alternating between radical and nucleophile chemistry. Indeed, the scientists were able to reconstruct the different steps of the reaction and propose a unique mechanism for Mmp10.
This study not only brings new insights into the complex process of methane production by bacteria, but also provides an unprecedented glimpse into how vitamin B12-dependent radical SAM enzymes catalyse chemically challenging reactions.
This study opens new routes for exploiting the catalytic potential of these enzymes for the development of new bioactive molecules and in green chemistry.
*Microbiome: community of microorganisms (bacteria, viruses, fungi, archaea) living in a given environment, e.g. in the soil, in the human digestive system...