Localization and quantification of antibiotics inside bacteria
Bacterial resistance to antibiotics is a major public health issue. The quantification of antibiotic accumulation is a crucial parameter that determines the antibiotics’ activity and is useful for the development of new antibacterial molecules. At the bacterial scale, UV fluorescence microscopy allows for tracking the kinetic accumulation of an antibiotic.
In this study, the MCT (Aix-Marseille University) and DISCO (Synchrotron SOLEIL) teams combined UV fluorescence microscopy results obtained at the DISCO beamline with cryo-X-ray fluorescence microscopy at the ID16A beamline (ESRF) to quantify antibiotic accumulation in isolated bacteria. These results open up a new avenue for studying metal-based antibiotics, which are receiving growing interest.
The activity of antibiotics depends on their ability to accumulate inside bacteria. Overproduction of efflux pumps (proteins located in the bacterial membrane, capable of exporting antibiotics out of the cell), coupled with the loss or reduction of porin production (proteins present in the outer membranes of certain bacteria, forming channels that allow antibiotic entry), often leads to a decrease in intracellular antibiotic concentration below a critical threshold for their activity, resulting in multidrug resistance bacterial phenotypes. In this context, quantifying antibiotic accumulation is crucial for guiding the development of new antibacterial molecules that limit recognition by efflux pumps and optimize their permeation through the outer membrane.
For several years, the MCT unit - Membranes and Therapeutic Targets - at Aix Marseille University has developed approaches based on the intrinsic fluorescence properties of certain antibiotics used in human therapy, such as fluoroquinolones. At the bacterial population level, spectrofluorometry provides a robust means to quantify antibiotic accumulation after different exposure times. At the single-bacterium level, UV fluorescence microscopy based on synchrotron radiation allows the tracking of antibiotic accumulation kinetics. However, this single-cell approach is limited by its spatial resolution of about 100 nm and the lack of fluorescence calibration, which prevents quantifying antibiotic concentrations in isolated bacteria. X-ray fluorescence (XRF) microscopy appears as a promising technique to overcome these limitations.
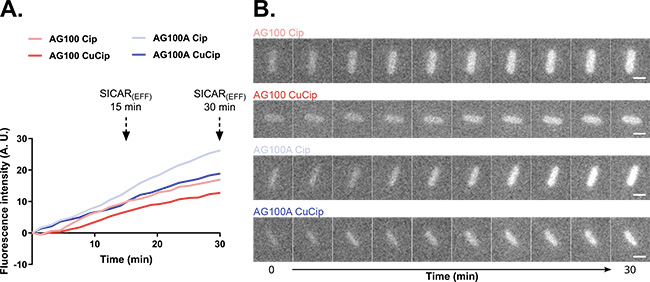
To quantify and localize antibiotic accumulation in isolated bacteria, a metal derivative of fluoroquinolone was used - the metal presence being detected by XRF. Its intracellular quantification in E. coli bacteria with or without efflux pumps provided results similar to those obtained by UV fluorescence techniques at the DISCO beamline using the "non-metallic" fluoroquinolone (Figure 1), validating the use of this derivative for this study. Moreover, using XRF under cryogenic conditions to quantify antibiotic accumulation allowed the visualization, at very high spatial resolution (20 nm), of antibiotic distribution within isolated bacteria (Figure 2). These results open a new path for studying metal-based molecules with antibacterial activity using XRF microscopy.
High-resolution spatial microscopy techniques are essential tools for the structural study of biological samples. The synchrotron approaches used each have their unique characteristics that allow the sample to be studied from different perspectives. Observing living cells with UV fluorescence microscopy or near-native state cells with cryo-SXT (Soft X-ray Tomography) and XRF microscopy offers a significant advantage: the absence of chemical treatments during sample preparation. The preservation of the structure and chemical composition of a sample is indeed crucial. Cryo-electron microscopy, which could not be used during this project, is also among the techniques that allow the study of biological samples close to their native state. The complementarity of these approaches enhances the overall understanding of the structure and composition of a sample.
Furthermore, synchrotron-based imaging techniques enable the tracking, quantification, and localization of antibiotic accumulation, which is necessary to understand and overcome the mechanisms behind the reduction in antibiotic accumulation and to combat multidrug resistance.