First structure of a rubella virus envelope protein
In the context of research on viral fusion proteins, researchers from the Structural Virology Unit at the Pasteur Institute, led by Felix Rey, have determined the first structure of a rubella virus protein. The envelope glycoprotein E1 is responsible for the binding of the virus to a cell surface receptor and for the membrane fusion of the virus with the target cell. The structure at 1.8 Å resolution, determined from data collected, notably on the PROXIMA 1 beamline, reveals the ectodomain of the E1 protein in its trimeric post-fusion form.
These results have been published in the journal Nature.
The rubella virus is the causative agent of German measles, which is usually a benign child disease. However, it can cause serious congenital birth defects in the fetus after maternal infection during pregnancy. While the use of the triple measles-mumps-rubella (MMR) vaccine has controlled rubella in developed countries during the last 40 years, congenital rubella syndrome remains a major public health problem in countries not using vaccine.
Nevertheless, congenital rubella syndrome continues to be an important problem in countries not using vaccine.
The rubella virus is the only member of the Rubivirus genus belonging to the Togaviridae family, which also includes the genus Alphavirus to which the chikungunya virus belongs. Despite its medical importance, little is known regarding the structural organization of the rubella virus.
Structure
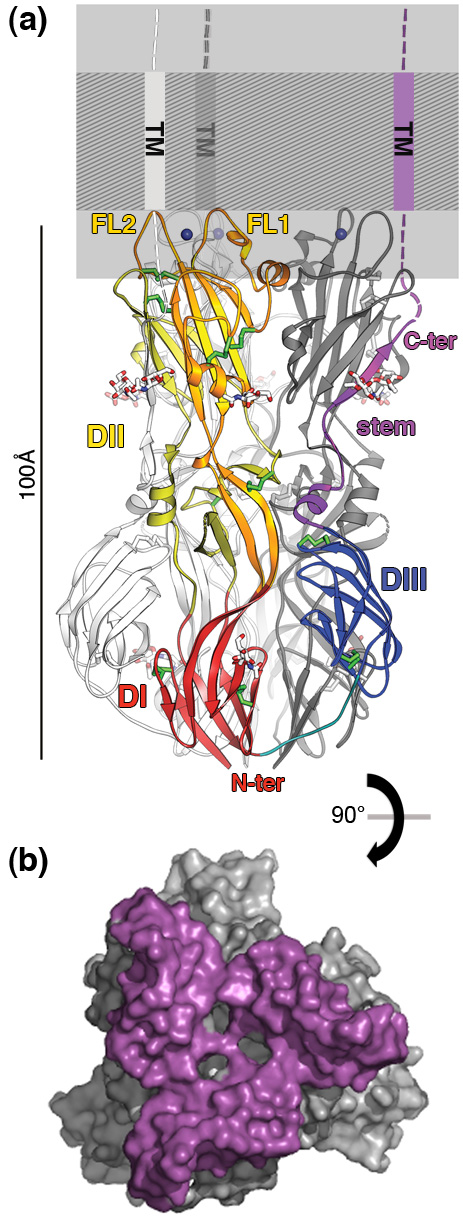
The E1 glycoprotein is the main antigen and the only target of neutralizing antibodies against rubella virus. Its structure has led to the localization of an epitope recognized by neutralizing antibodies, as well as allowing the neutralization mechanism to be identified. E1 is also involved in target cells entry carrying both the receptor-binding and the membrane-fusion functions. The structure of the E1 trimer reveals a very large membrane-fusion surface composed of two loops and one metal-binding site per protomer (see Figure). This feature is unique when compared to the membrane fusion envelope proteins of alphaviruses and flaviviruses. The extent of the fusion surface suggests that it could induce significant pressure during membrane insertion of the virus in order to catalyze more effectively the fusion of viral and cell membranes.
The metal binding sites observed in the E1 trimer can be occupied by a sodium or calcium ion, and the binding properties of E1 to lipids are different according to the nature of this ion and pH.
The presence of a metal in E1 evokes the cell proteins of the "T-cell immunoglobulin and mucin (TIM)" family (2), which specifically recognize, via a metal, phosphatidylserine lipid head groups on the cell plasma membrane in apoptosis (programmed cell death).
Evolution
The rubella virus E1 protein is a class II membrane fusion protein, a class that, until now, has included the envelope proteins of alphaviruses (3), and flaviviruses (4). These latter viruses have arthropods as vectors (mosquitoes, ticks, etc.), unlike the viruses of rubella, measles and mumps, in which inter-human transmission is via airborne droplets. Structural comparisons show that the class II fusion proteins from alphaviruses and flaviviruses are structurally closer to each other than they are to rubella virus E1 protein.
This observation suggests that repeated cycles of transmission between vertebrates and arthropods imposes constraints that result in more conservative evolution for their fusion proteins. In the absence of this constraint associated with viral replication in the different hosts, the strictly human rubella virus has evolved towards a unique niche as the only member of the Rubivirus genus.
.
(b) Surface accessible to the solvent in the E1 trimer (90° to (a)), showing the extent of the fusion surface viewed from the membrane (~8000 Å2, in purple).
References:
1. Wolinsky JS, Sukholutsky E, Moore WT, Lovett A, McCarthy M, Adame B. An antibody and synthetic peptide defined rubella virus E1 glycoprotein neutralization domain. J. Virol. 67, 961–968 (1993).
2. Santiago C, Ballesteros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casasnovas JM. Structures of T cell immunoglobulin mucin receptors 1 and 2 reveal mechanisms for regulation of immune responses by the TIM receptor family. Immunity 26, 299–310 (2007).
3. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. Nature 427, 320-325 (2004).
4. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Heinz FX, Rey FA. EMBO J. 23:728-738 (2004).